Concepts¶
Names and taxa¶
Groups of related organisms are identified by scientific names, but many taxa have more than one name. Taxonome treats synonyms as equivalent references to a single taxon object. In addition, each taxon has one accepted name (its name property). Taxonome does not force you to use any particular taxonomy: the accepted name is simply the name which your data source treats as accepted.
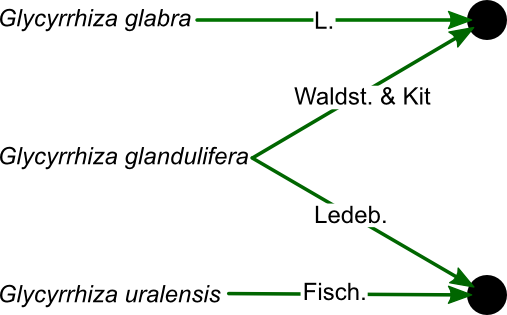
In some cases, the same name may have been given to different taxa (homonyms). Rigorous sources will give all names with an author citation. The combination of a name and authority is encapsulated in Taxonome’s Name class, and we’ll refer to it as a “qualified name”. Taxonome assumes that a qualified name uniquely identifies one taxon.
For example, two species, each with an accepted name, and a homonym which can refer to either, can be thought of like this:
Collections of taxa¶
Taxonome works with datasets which represent collections of taxa; for example, ‘Grass species from the GRIN database’. A dataset stores species by scientific name, and is stored to allow matching with synonyms or small spelling variations. Each taxon can also store other information, such as distribution or common names.
Web services for taxonomic data are represented in taxonome in a similar way. When you search them, taxonome make a web request, and processes the results to a common format. For example, taxonome.services.tropicos is a module which queries the Tropicos API for plant taxa.
The technical detail: Standard datasets are instances of TaxonSet, which inherits from the abstract class TaxaResource.
Distribution¶
A taxon has a distribution parameter, which can be a set of the regions where it is known to occur. The module taxonome.regions contains tools for working with these. See Distribution data for more information.