Using the application¶
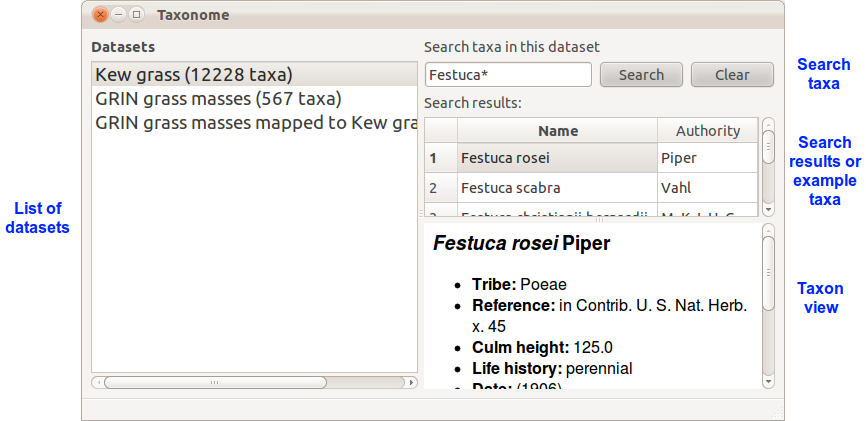
The list of datasets on the left shows the datasets you have loaded. Click on a dataset to select it, or double click to rename it.
With a dataset selected, you can use the search box on the top right. It accepts species names or wildcard searches, and searches synonyms as well as accepted names. “Festuca*” will find any species which the dataset lists in the genus Festuca, including species with a synonym in Festuca. The results are displayed (by accepted name) in the panel below. When no search is active, the same panel shows 10 example taxa from the selected dataset.
The taxon view panel in the bottom right shows a taxon that has been selected in the search results/example taxa panel. It displays details loaded from the dataset, such as synonyms or distribution.
Loading datasets¶
From the file menu, you can load datasets from CSV files, which are available from many sources online. A dialog will ask you to select which columns contain the species names and authorities. There are three different types of data that you can load from CSV files:
- Taxa: Each row refers to one taxon.
- Synonyms: Each row connects one name to the accepted name for that taxon.
- Individual records: Each row refers to a separate specimen, sighting, population, or something similar that is assigned to a taxon. The same name may occur on many rows.
You can also save and load data in a format specific to Taxonome, called JSONlines in the menu.
Taxonome can also fetch datasets over the web (‘Fetch collection of taxa’ in the web services menu). At present, this only works with the USDA GRIN database, but we hope to add more services in the future.
Matching names¶
The core feature of Taxonome is matching species names to a given set of names, accounting for synonyms and spelling differences. To start, you need to have a dataset loaded for the target synonymy you want to use. For example, you may download synonym data for a family from GRIN, as described above. Or see Reading specific data sources for scripts to load some other databases.
Then, from the Taxa menu, select Match taxa by name. Ensure that the Match to names in option points to your target synonymy. You have the option to match taxa from a loaded dataset, or from a CSV file. Matching directly from a CSV file is useful if you have a very large dataset: Taxonome can read it row-by-row, rather than loading the whole file into memory. If you pick a CSV file, the next screen will ask you to select the name and authority columns, just like when loading a dataset from CSV.
The next screen has a number of options to control the name-matching process:
- How should subspecies be treated if the target synonymy does not contain a matching subspecies? By default, only nominal subspecies (e.g. Zea mays subsp. mays) can be matched to their parent species, but you can choose to reject all unmatched subspecies, or match them all to a parent species.
- Where there are multiple matching names (homonyms), the one (if it exists) which the target synonymy considers an accepted name is typically the most likely match. By default, Taxonome will select this automatically when matching a plain name without an authority. It can be set to always automatically pick the accepted name, or never to do so.
- If a name only has one match, but the authority for the name does not match that for the taxon, Taxonome will normally allow the match; this can be turned off.
- When a name has multiple matches, Taxonome can present you with the matches to make a manual selection. This is off by default, but can be enabled here. How many times it asks you to choose will depend on your data, but as an example, matching some 700 grass names from GBIF produced about 10 questions to answer manually.
The final screen offers a number of options for output from the matching process.
- Taxa data with new names will save the input taxa data (as CSV) along with the names they were matched to. Taxa that could not be matched are not included.
- Name mappings saves a CSV file of the input names matched to the target names, including input names which weren’t matched (with empty fields for the target name).
- Full log of matching process saves a CSV file detailing the steps used to match each name.
- You can also choose to make a Taxonome dataset of the input taxa with the matched names. This will store the taxa in memory, so you shouldn’t use it with very large (multi-gigabyte) datasets.
Checking the matched names¶
You should check the matches Taxonome finds. Turn on the Name mappings output, and when the matching has finished, load the CSV file in a spreadsheet application. The first two colums show the original name and authority, the next two show the matched name and authority, and the fifth column shows the closeness of any fuzzy matching done. The lowest numbers here are the least certain matches, so you can sort using this column to find them.
If the matched name looks very different from the input, it has probably been matched through a synonym.

To get more detail about the steps Taxonome used, you can turn on the Full log of matching process output. This produces a CSV file with several rows for each input name, showing intermediate steps like matched synonyms.
Combining datasets¶
When you have a number of datasets loaded using a common set of names, such as after matching them to a preferred synonymy, you can combine them. From the Taxa menu, select ‘Combine datasets’.
You can add source datasets to one of two groups: ‘target’ datasets and ‘background’ datasets. A taxon is included in the results if it is in any target dataset, with information attached from all the datasets in use which have the same taxon. Background datasets are used to add information, but their taxa are not automatically included.
For species’ distribution information, you can choose to get distribution data from one dataset, or to combine distribution information from all the datasets in use.